MINDS: Multimodal Integration of Oncology Data System
Integrating Disparate Multimodal Datasets
Aakash Tripathi
Department of Machine Learning, Moffitt Cancer Center & Research Institute,
Electrical Engineering Department, University of South Florida.
Asim Waqas
Department of Machine Learning, Moffitt Cancer Center & Research Institute,
Electrical Engineering Department, University of South Florida.
Ghulam Rasool
Departments of Machine Learning and Neuro-Oncology, Moffitt Cancer Center & Research Institute,
Electrical Engineering Department and Morsani College of Medicine, University of South Florida.
[Journal Paper] [Code]
______________________________________________________________________________________________________
Overview
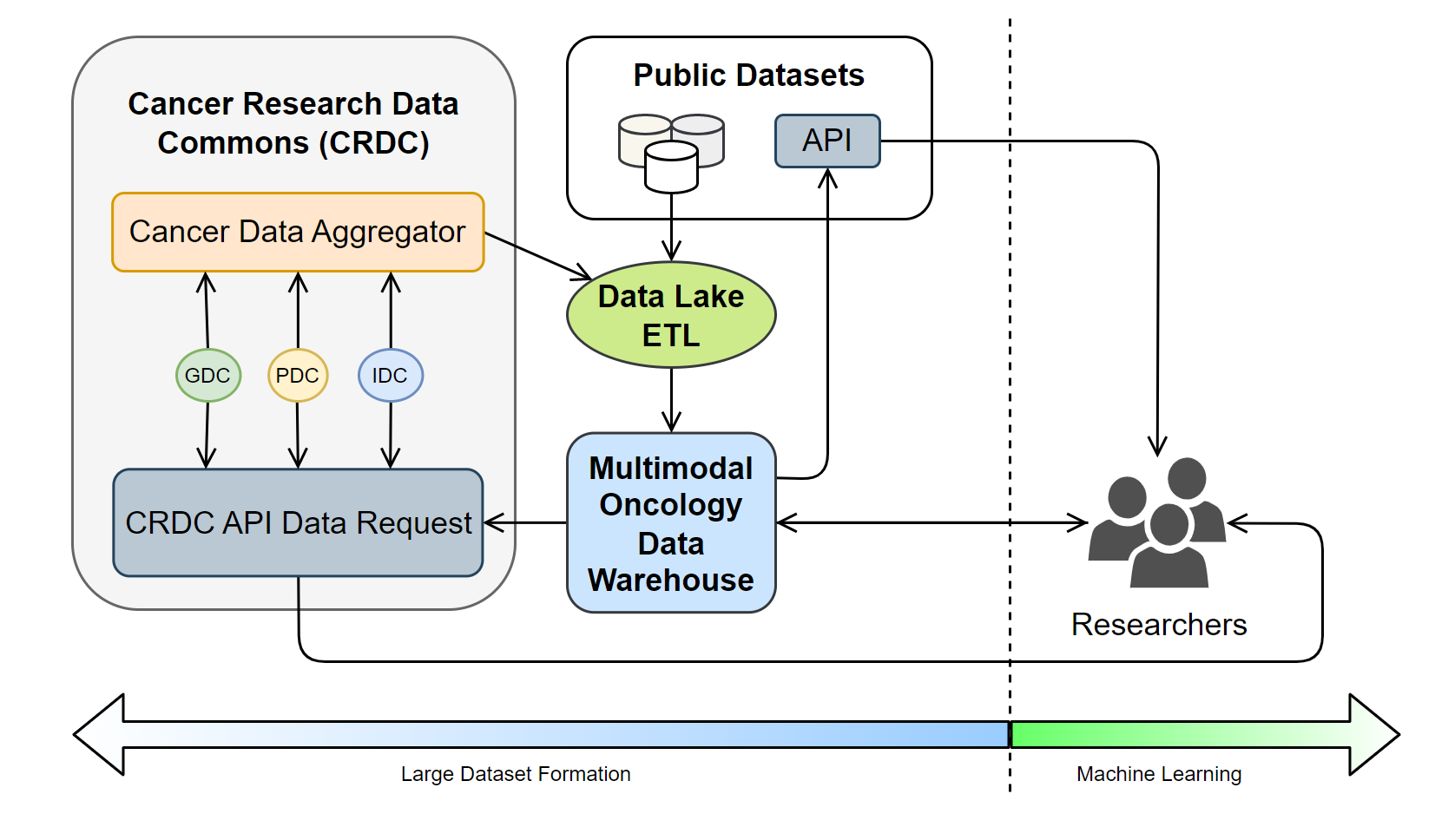
The rapid growth of high-throughput technologies and healthcare digitization has led to a surge in oncology data, providing unprecedented opportunities for cancer research and the development of tailored treatments. Nevertheless, such data's voluminous and multimodal nature poses substantial storage, management, and analysis challenges. Traditional multimodal data is collected from clinical trials, which undergoes a meticulous standardization and quality control process and is subsequently stored in biobanks. However, this approach results in fragmented data that fails to capture a comprehensive health status. To overcome this, the National Cancer Institute (NCI) developed the Cancer Research Data Commons (CRDC), a cloud-based infrastructure that unifies various data repositories. Furthering this effort, the Cancer Data Aggregator (CDA) was introduced to harmonize and consolidate data across the CRDC. Despite these advancements, fragmentation remains at the modality level, especially when dealing with data across biobanks. To resolve this, we propose a novel system, a data lakehouse architecture in the cloud, specifically leveraging Amazon Web Services (AWS), uniquely designed to manage and integrate vast, multimodal oncology data efficiently. This innovative system does not store large amounts of unstructured data but maps metadata about patients' unstructured data to a data warehouse. This mapping allows us to pull data from the biobanks only when queried, offering a solution to the data fragmentation issue prevalent in current systems. As a result, we can achieve a streamlined data management process while maintaining the integrity and security of the patient's sensitive health information.
______________________________________________________________________________________________________
Method
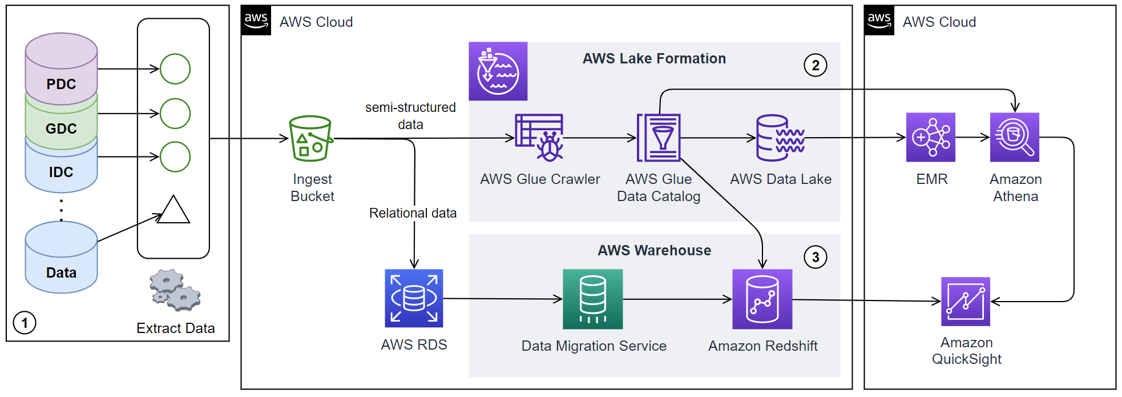
Our implementation consists of a three-stage process: data acquisition, warehousing, and consumption.
- The data acquisition stage involves gathering raw structured and semi-structured data and metadata about unstructured data from sources such as Genomics Data Commons (GDC), Proteomics Data Commons (PDC), and Imaging Data Commons (IDC), storing them in a data lake.
- The data is then extracted, transformed into a more structured format, and loaded into the data warehouse using ETL (Extract, Transform, Load) tools.
- Finally, the structured data in the data warehouse is utilized for various purposes, including feeding into analytics dashboards for visual representation or providing data for machine learning models.
We utilize several AWS tools to implement this methodology. Initially, all publicly available structured and semi-structured data from the data commons portals are gathered, and the semi-structured data is uploaded into an Amazon S3 ingest bucket, while the structured data is stored in an AWS Relational Database Service (RDS) instance. The data is then cleaned, preprocessed, and aggregated using Amazon AWS Glue. The AWS Data Lake Formation tool transforms the semi-structured data stored in the S3 bucket into a queryable data lake. The structured data stored in the AWS RDS is integrated into a data warehouse using Amazon Redshift. Our system gives users a dashboard built on Amazon QuickSight, enabling data visualization and interactive analysis. Users can select a cohort of cases based on their research needs, and our system will generate the corresponding structured data for training machine learning models using AWS Step Functions, which coordinate and manage the data export process. The AWS Glue crawler is used in data extraction, transformation, and loading (ETL). It establishes secure connections to databases, uses custom classifiers to catalog the data lake, and employs built-in classifiers for ETL tasks. It then merges catalog tables into a single database and uploads the catalog to a data store. AWS Lambda triggers the AWS Glue crawler whenever new data is added to the S3 bucket. The data warehousing stage uses Amazon Redshift, designed for high-performance analysis and reporting of large datasets. The structured data from the RDS instance is loaded into Redshift using AWS Glue ETL jobs. The data consumption stage is facilitated by Amazon QuickSight, which enables data visualization and interactive analysis. The entire system is orchestrated using AWS Step Functions, which lets you coordinate multiple AWS services into serverless workflows.
To ensure the security and privacy of the data, we employ several AWS security services and best practices. Data is encrypted at rest using AWS Key Management Service (KMS) and in transit using SSL. Access to the data is managed through bucket policies, access control lists, IAM policies, and AWS Lake Formation. AWS Glue provides a secure, isolated environment for ETL jobs. Amazon QuickSight uses AWS IAM and AWS Lake Formation for access control. Monitoring and logging are accomplished using AWS CloudTrail and Amazon CloudWatch. We use AWS Auto Scaling to automatically adjust capacity and Amazon RDS Multi-AZ deployments for high availability and failover support for DB instances to ensure the system's reliability and availability.
______________________________________________________________________________________________________
Conclusion
The successful implementation of our proposed solution will result in an efficient, integrated system for managing large volumes of multimodal oncology data. By leveraging the capabilities of AWS and a data lakehouse architecture, the system will overcome the current challenges of data fragmentation and inefficiency in data retrieval. This solution will extract pertinent information from vast, heterogeneous clinical data repositories, efficiently streamlining data management processes. With its ability to work with extensive datasets, the system promises to improve data accessibility and utility, potentially enhancing the accuracy of cancer research and the development of personalized treatments. The solution's high security and reliability ensure data integrity and privacy, thus demonstrating a potential to significantly improve current data management practices in oncology research.
______________________________________________________________________________________________________
Citation
@Article{minds,
AUTHOR = {Tripathi, Aakash and Waqas, Asim and Venkatesan, Kavya and Yilmaz, Yasin and Rasool, Ghulam},
TITLE = {Building Flexible, Scalable, and Machine Learning-Ready Multimodal Oncology Datasets},
JOURNAL = {Sensors},
VOLUME = {24},
YEAR = {2024},
NUMBER = {5},
ARTICLE-NUMBER = {1634},
URL = {https://www.mdpi.com/1424-8220/24/5/1634},
ISSN = {1424-8220},
DOI = {10.3390/s24051634}
}
