Cells have energetic and anabolic needs for growth and proliferation. Our research is focused on understanding how cells meet these needs and the mechanisms governing the regulation of tumor metabolism in vivo. While cell culture systems are useful for querying the activity of metabolic pathways, many factors in vivo likely play a major role in the regulation of cellular metabolism, including tumor/stroma interactions, contact with the extracellular matrix, local hypoxia and nutrient availability and nutrient intake by the host. The DeNicola laboratory investigates the influence of both genomic alterations and the microenvironment on cellular metabolism in vivo.
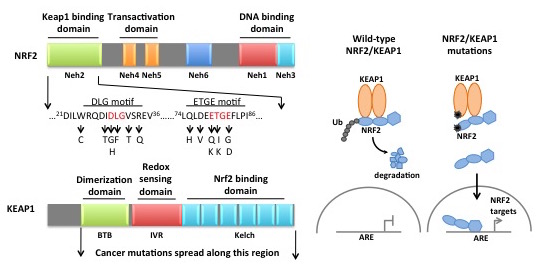
One of the most commonly mutated pathways in NSCLC is the NRF2/KEAP1 circuit. NRF2 is a stress-responsive transcription factor that directs select transcriptional programs in response to oxidative stimuli. KEAP1 directs NRF2 destruction. NRF2 and KEAP1 mutations lead to loss of NRF2 degradation and constitutive NRF2 accumulation, thereby promoting glutathione synthesis, detoxification of reactive oxygen species (ROS) and proliferation. To assess the effects of NRF2 activation on cellular metabolism and tumorigenesis in an isogenic system, we generated genetically engineered mouse models of both the NRF2D29H and KEAP1R554Q mutations found in human NSCLC. Our laboratory is currently characterizing these models.
Mutations in KEAP1 and NRF2 lead to metabolic deregulation. Despite the identification of the first KEAP1 mutations in 2006, the precise mechanisms by which NRF2 promotes tumorigenesis are unclear. We, and others, have identified that NRF2 activation leads to profound deregulation of cellular metabolism, including the promotion of serine biosynthesis and cysteine metabolism. We are currently evaluating both the tumor promoting and antagonistic metabolic processes regulated by NRF2. Further, we are evaluating NRF2-regulated metabolism in pancreatic cancer, which we previously found has high activity of NRF2 due to signaling downstream of oncogenic KRAS. A major research focus of the DeNicola lab is investigating how NRF2 promotes tumorigenesis, metabolic deregulation, and metabolic dependencies in vivo using genetically engineered mouse model systems.
Our laboratory utilizes a combination of gas chromatography mass spectrometry (GC-MS), targeted liquid chromatography mass spectrometry (LC-MS/MS, with triple quadrupole) and non-targeted LC-MS (with orbitrap) to profile and quantify metabolite changes in cells, fluids and tissues. With these technologies we perform both metabolite quantitation and flux analysis using stable isotope labeled substrates. We are currently focused on sulfur metabolism, particularly sulfur-containing metabolites downstream of cysteine.