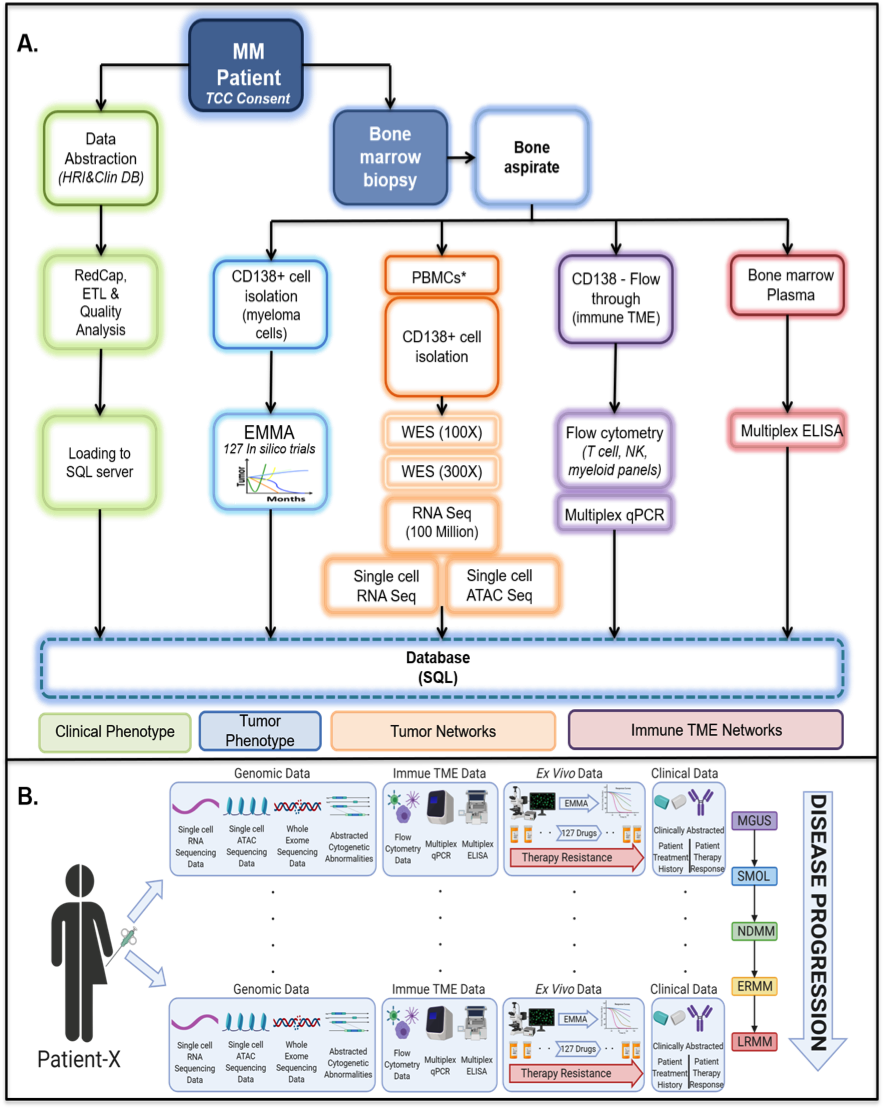
An overview of the multi-omics database developed by our group to understand patient-specific drivers of Multiple Myeloma. A, An overview of the hierarchical structure of the data and flow of samples and information collected from patients and biopsies. B, A graphical visualization of myriad aspects of the data linked to a single patient across various disease states accessible via the multi-omics relational database.
As a founding member of Oncology Research Information Exchange Network (ORIEN), M2GEN and the Moffitt Cancer Center have collected longitudinal clinical data, tissue samples, and molecular data from 5,337 different samples of cancer patients along 10 years through the Total Cancer Care (TCC) protocol. Importantly, our multi-disciplinary team of investigators has led the Moffitt MM Working Group (MMWG) in the establishment of infrastructure to consent MM patients, collect extra bone marrow aspirate for research protocols, positively select for MM cells, and submit them for bulk sequencing (RNAseq and WES) through ORIEN/AVATAR project. To date, we have collected 1,123 different MM samples, which are sequenced in batches of 12-15 specimens on a monthly basis; thus, accruing patients and collect samples at a rate of ~160/year. Our data repository currently consists of 844 RNAseq and 870 WES individual samples. In addition, clinical data and treatment history of these patients are abstracted from electronic health records (EHR). The final components for our study are phenotypic characterization of these patient specimens with an ex vivo drug sensitivity assay EMMA and multiparameter flow cytometry (MPF) on lymphocyte and myeloid populations. EMMA is a high-throughput ex vivo drug sensitivity assay of primary MM cells in a reconstruction of the bone marrow microenvironment. To date, we have tested 537 primary MM samples in EMMA. Using MPF we are able to characterize bone marrow T cell populations and myeloid populations. We have used this comprehensive cohort to investigate the underlying mechanisms driving MM progression and evolution of drug resistance in both tumor and iTME.
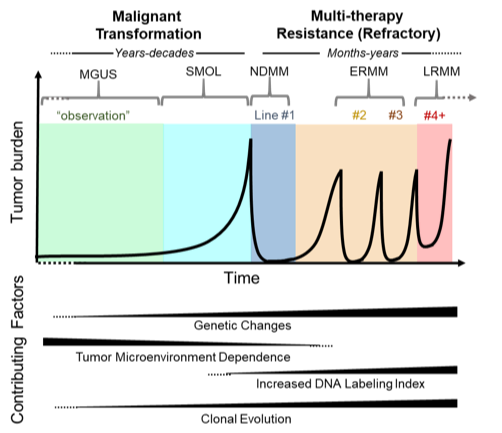
We propose that transition from pre-malignant - monoclonal gammopathy of undetermined significance (MGUS) or smoldering MM (SMOL) - to active disease (newly diagnosed, NDMM) consists of loss of bone marrow microenvironment dependency and accumulation of genetic abnormalities.Successive lines of therapy response are followed by early relapses (ERMM, 1-3 lines of therapy) and eventually refractory disease (late relapse MM, LRMM, >3 lines of therapy), characterized by high expression of cell cycle genes and increased genetic heterogeneity.While MM progression is selected by microenvironment-imposed restrictions (i.e., limited availability of plasma cell-supporting niches in the marrow), refractory disease is the product of selection for the fastest growing therapy-refractory clones during relapse and tumor re-growth.
We have explored the central biology involved in MM progression and refractory disease by leveraging a new database of combined clinical and molecular data, from a cohort of 1,016 bone marrow aspirates collected from patients across MM spectrum treated at Moffitt Cancer Center, and identified mutations, cytogenetic events and epigenetic dysregulation driving MM progression and emergence of multi-drug resistance.
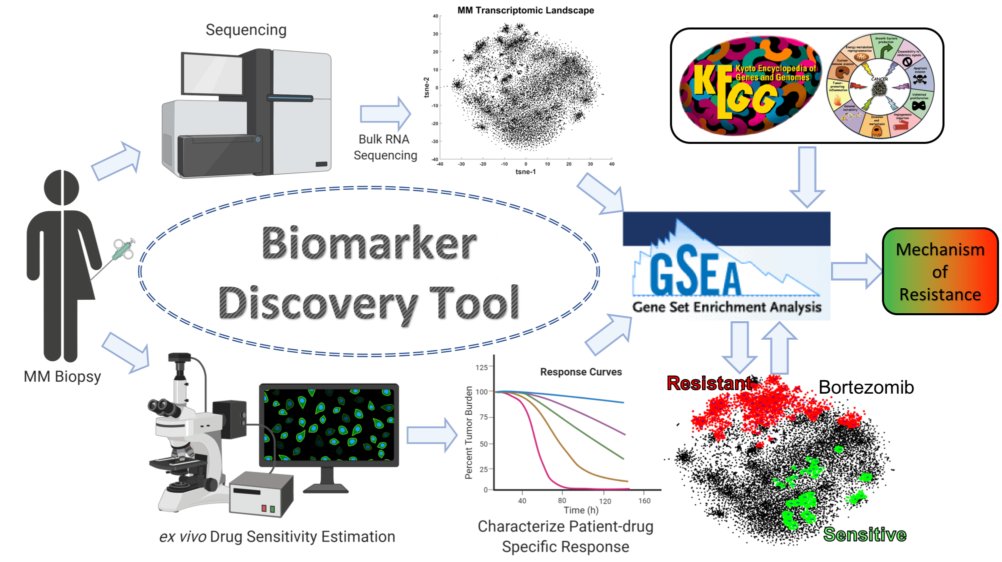
To date, we have tested drug sensitivity of over 400 patient-derived Multiple Myeloma samples with standard-of-care and experimental drugs. In this project, we investigate mechanisms of resistance to therapy in MM, with two purposes: first, to develop biomarkers to identify the ideal regimen of each patient, and second, to propose new combination therapies capable of reversing drug resistance.
Our approach consists of using RNA-seq data to construct a MM transcriptomic topology, by breaking down the complexity of transcription regulation of tens of thousands of genes by grouping them in clusters of co-expressing genes, with putative common mechanism of regulation.
By correlating the expression of these gene clusters with ex vivo drug sensitivity of matching bone marrow biopsy MM samples, we identify clusters of genes that are enriched for drug resistance (in red) or sensitivity (in green). The clusters implicated in resistance, or sensitivity are studied to find associations with KEGG pathways and Cancer HALLMARKS that provide evidence into mechanism of resistance.
ClinicalTrials.gov Identifier: NCT04151667
Detailed Description: In this response adapted approach, older adults with newly diagnosed symptomatic multiple myeloma will receive daratumumab and dexamethasone for 2 months. Patients who achieve a partial response or better will continue on daratumumab. Patients who achieve less than a partial response will have lenalidomide or bortezomib added to their therapy. Patients who experience progressive disease on daratumumab after the initial 2 months of monotherapy or on the combination of daratumumab and either lenalidomide or bortezomib will come off study.
Additionally, as a secondary exploratory aim, EMMA is used to assess drug sensitivity of primary MM cells from bone marrow biopsies collected at trial enrollment, after two cycles of therapy, and relapse, to identify the most promising agent to be added to daratumumab, and estimate depth and duration of response.
Coming soon...