Sleeping Beauty insertional mutagenesis is a gene discovery tool
Sleeping Beauty (SB) is a gene discovery platform that utilizes a DNA transposable element engineered to mutagenize the mouse genome through random genomic integration and selection. SB transposon insertions perturb regulation of cancer-causing genes. The system has been utilized to model human hematopoietic and solid tumor types. Forward-genetic screens using SB in the context of initiating oncogenic events have identified cooperating cancer drivers that promote tumorigenesis. SB is a powerful tool for comparative genomic analysis of human cancers to prioritize cancer causing genes for functional studies.
The Sleeping Beauty (SB) transposon was reverse-engineered from salmonoid fish to construct a DNA-based insertional mutagen that could be introduced into the mouse genome as a transgene and mobilized by the SB transposase, expressed in trans. The transposase recognizes and binds to the inverted repeats at the ends of the transposon, initiates a double-strand break (DSB), and liberates the transposon from the donor concatemer or an integration site elsewhere in the genome. The mobilized transposon can then integrate anywhere in the genome at a TA dinucleotide.
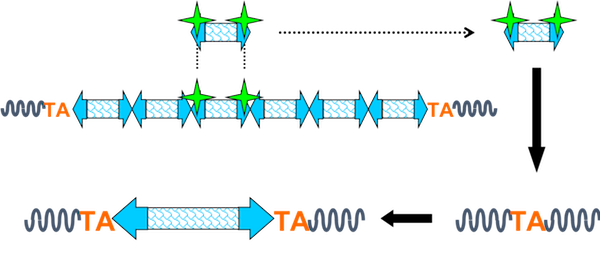
The transposon contains a minimal internal promoter and a splice donor, such that it can drive expression of downstream exons when integrated into the sense direction of the coding gene. Here, the murine stem cell virus promoter is shown. Other transposons have been engineered to contain the CAG promoter. The transposon also contains a splice acceptor site and a bidirectional polyA tail, such that it can act as a gene trap and disrupt gene expression. An inducible allele encoding the transposase allows for the control of transposition initiation in space and time.
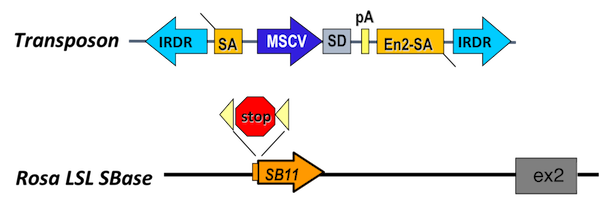
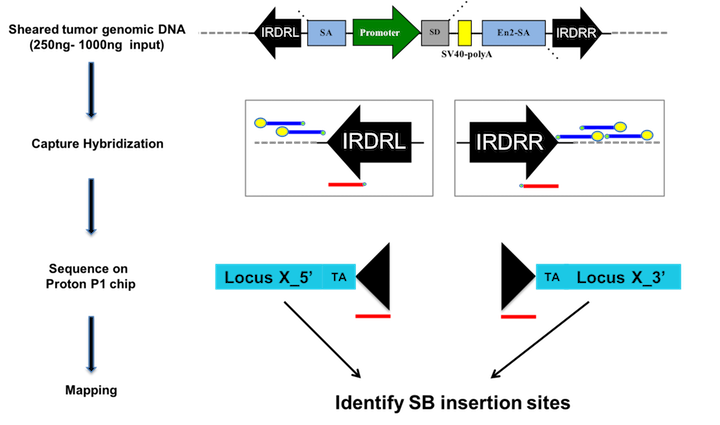
We can combine SB with mutations that confer cancer predisposition to a cell type of interest or provide the first "hit" in the development of cancer. Inducible mutations and the SB transposase are activated concurrently upon Cre recombinase expression driven by a tissue specific promoter of choice. For the SB PDAC model, we combined SB with the KC mouse model to interrogate genes that cooperate with oncogenic KrasG12D to drive tumor progression. Tumors are harvested at study endpoint and histologically confirmed. Genomic DNA is extracted from tumors for sequencing SB insertion sites in individual tumors. Drivers that cooperate with oncogenic KrasG12D are identified in the tumor cohort by statistical analysis of genes with more insertions than expected by chance at the population level.

We recently developed a semi-quantitative probe-based sequencing method to detect transposon insertions from bulk tumor down to single cell resolution (Mann et al. Nature Biotech 2016). We use capture hybridization with probes directed against the IRDRs (red) to enrich for sequences containing the transposon, while blocking enrichment of unmobilized transposon (blue probes). By sequencing the DNA we can identify clonal insertions in an individual tumor based on genomic mapping and sequencing read-depth. Statistical analysis at the population level identifies recurrently mutated driver genes. SB insertion patterns across tumors can be utilized to identify co-occurring drivers that may cooperate during tumorigenesis. Single cell sequencing defines cooperating drivers in sub clones of individual tumors.