Myeloid leukemia is a molecularly heterogeneous disease. While specific chromosomal aberrations have been defined in a subset of patients, the majority of adult acute myeloid leukemia (AML) is characterized as chromosomal normal (CN-AML). Current research efforts are focused on identifying new therapies to treat this disease. However, the paucity of major molecular aberrations identified for CN-AML presents a challenge for identifying specific therapeutic targets and predicting therapeutic outcome. We utilized Sleeping Beauty to model a completely penetrant, highly aggressive chromosomal normal (Cn) myeloid leukemia (SB_ML). Statistical analysis of transposon insertions defined a number of oncogenic drivers; human homologs of the SB_ML drivers are enriched for mutations in human AML, and both human AML and SB_ML drivers are significantly enriched in JAK-STAT and MAP kinase signaling.
Using single cell sequencing, we showed that ML is characterized by a high degree of intratumor heterogeneity that was not appreciated from bulk tumor sequencing and identified several cooperating relationships in an individual tumor between the most frequently mutated drivers, validating their biological impact in driving ML (K Mann et al. Nature Biotech 2016). 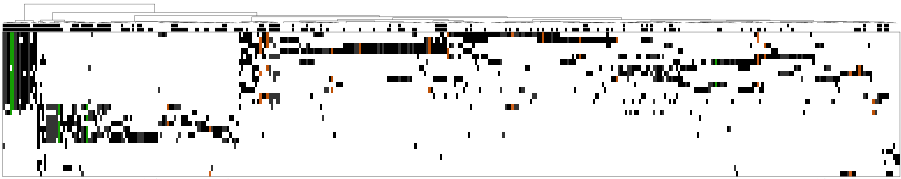
We are exploiting this intra-tumor heterogeneity to characterize cooperating sub clones that drive tumor progression using purifying selection in transplantation models. We can then utilize this information to predict which genetic aberrations contribute to cancer progression and potentially therapeutic resistance.
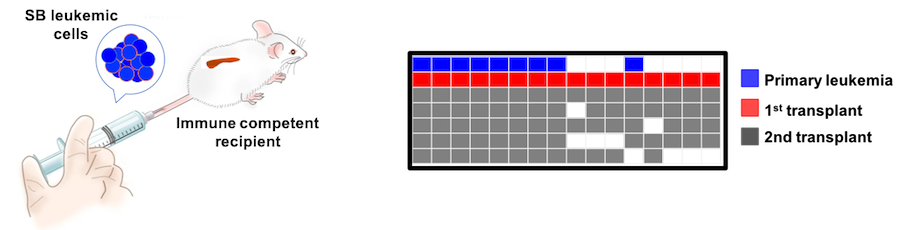
The SB_ML model is advantageous for therapeutic studies in that the inherent mutation burden and ITH is more representative of the molecular landscape of human AML than are other GEMM models of AML, which typically rely on the presence of a specific sensitizing mutation or expression of a translocation product to drive disease. Using SBCapture Seq, we can determine in a quantitative manner which insertions are lost, gained or maintained upon therapeutic intervention and characterize the effect of treatment on remodeling tumor heterogeneity.
