Wang Lab: developing new methods for cancer biomarker discovery
Machine Learning methods for cancer outcomes
The primary methodology interest in our group is to develop efficient and effective machine learning methods (including kernel machine, gradient tree boosting, and deep learning) for biomarker discovery. We are particularly interested in making state-of-the-art machine learning methods available for survival outcomes, which are primary outcomes in cancer research. Below is an example of extending the extreme gradient tree boosting (XGBoost) method to fit the nonlinear (Gaussian kernel) risk in the cox model.
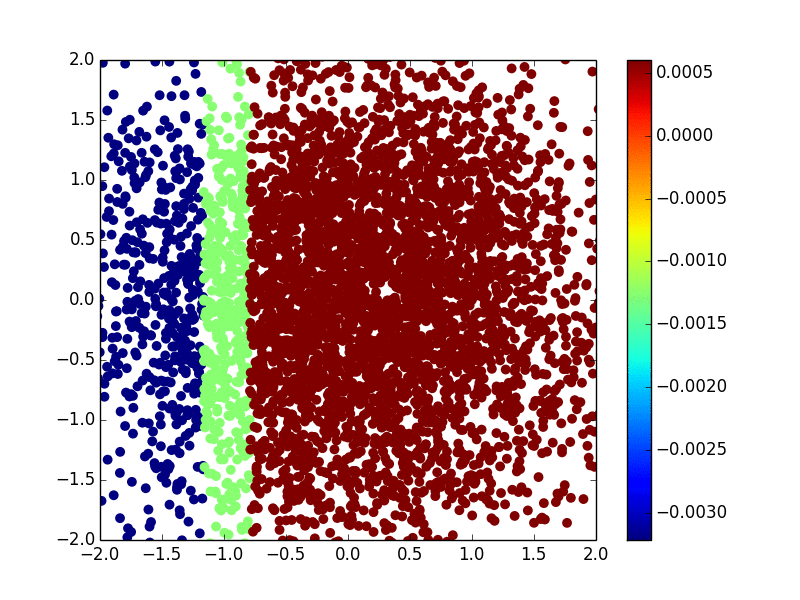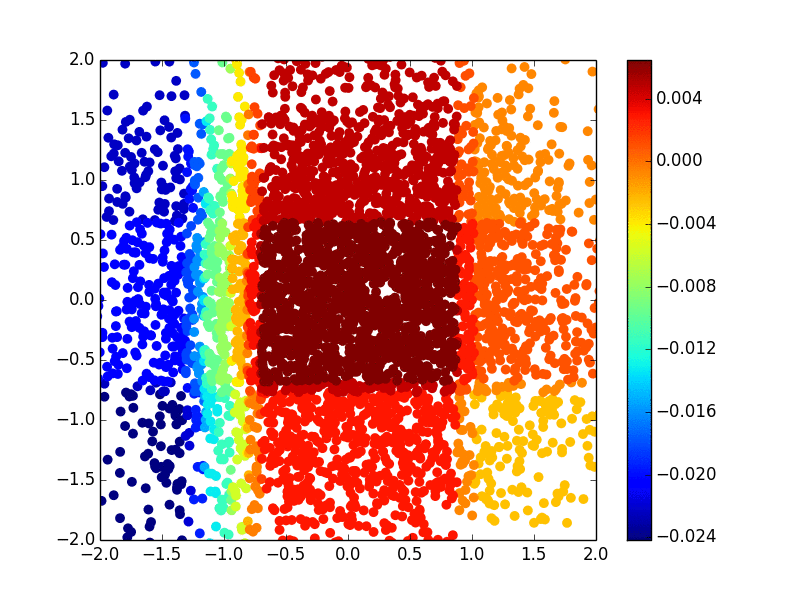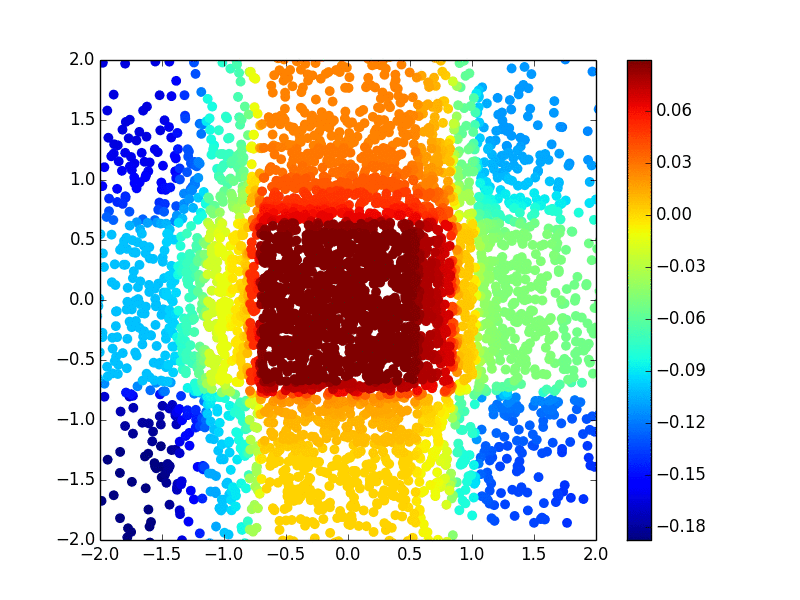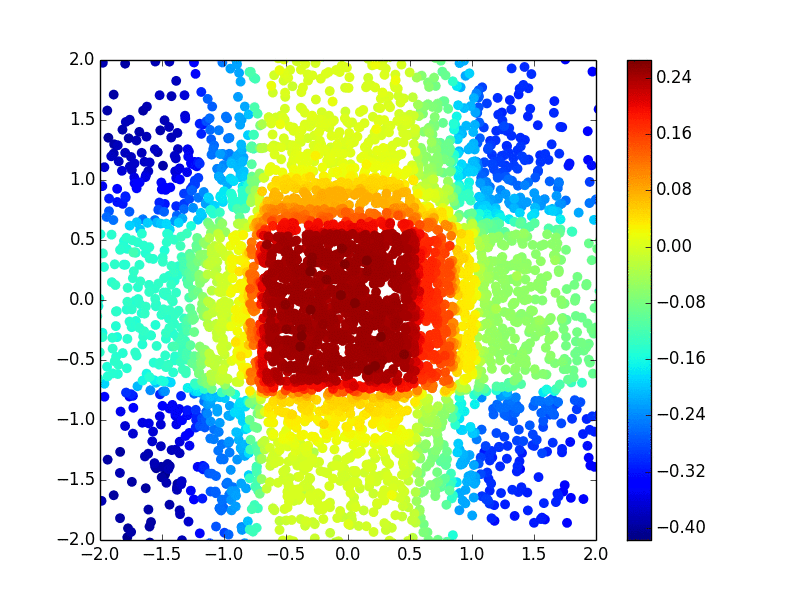
Tools and methods for CNV, methylation and ncRNA
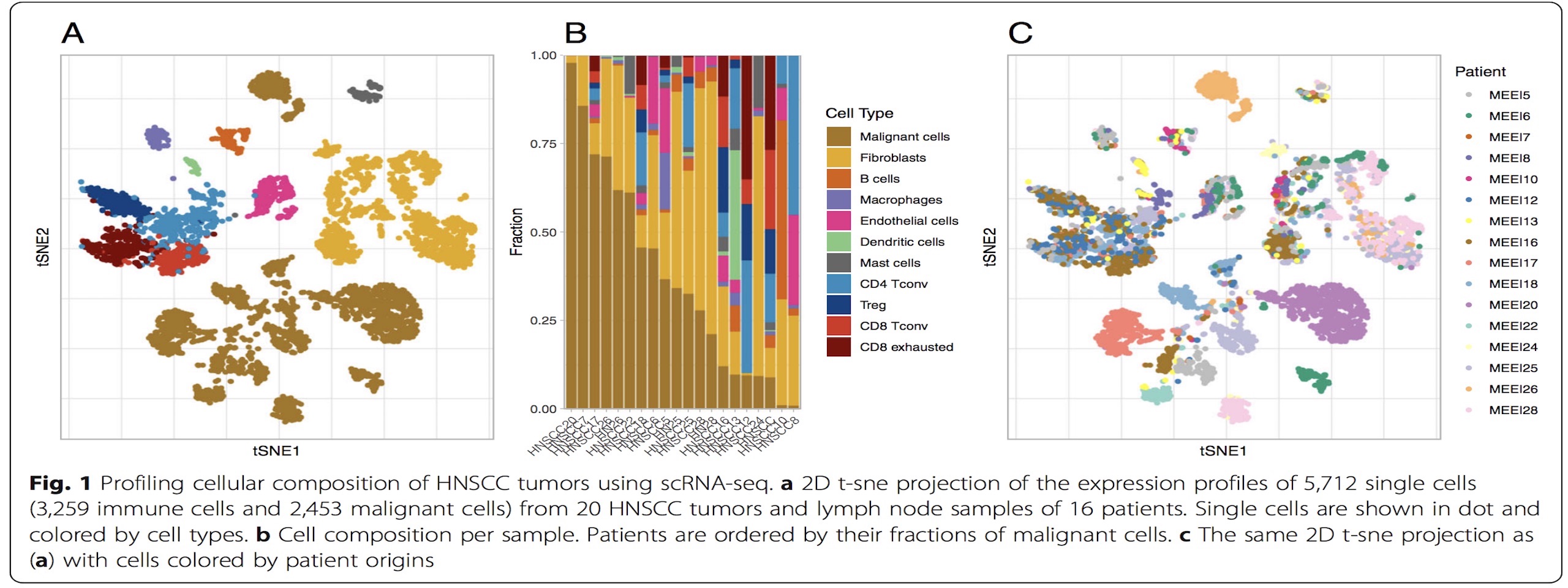
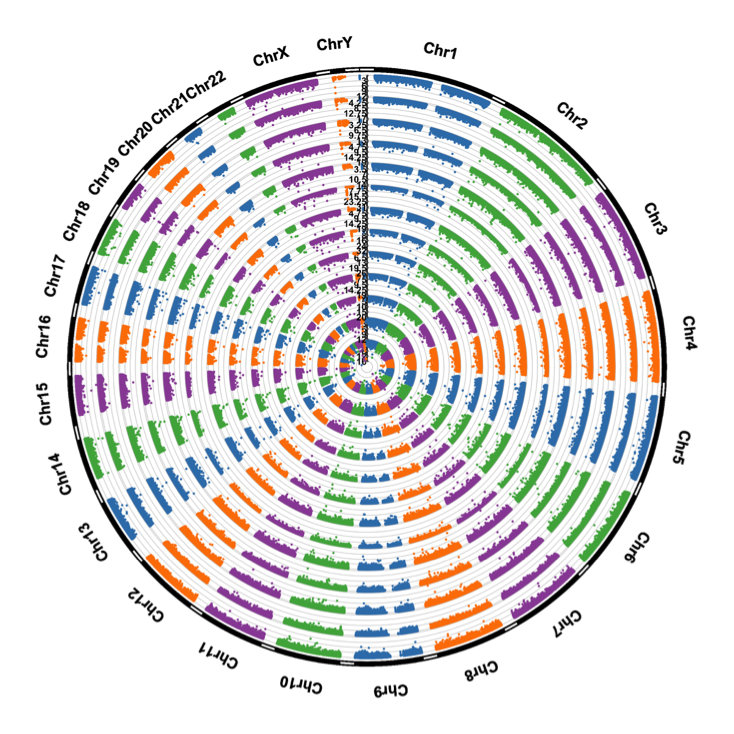
Our group is interested in developing novel statistical and computational tools for selecting predictive and functional biomarkers from less-investigated yet informative Omics data including copy number variations, methylation data and non-coding RNA. We have successfully applied our methods to perform genome-wide prognostic biomarkers based on CNV (as shown in the figure below) and CpG biomarkers (https://tcga.shinyapps.io/imme/)

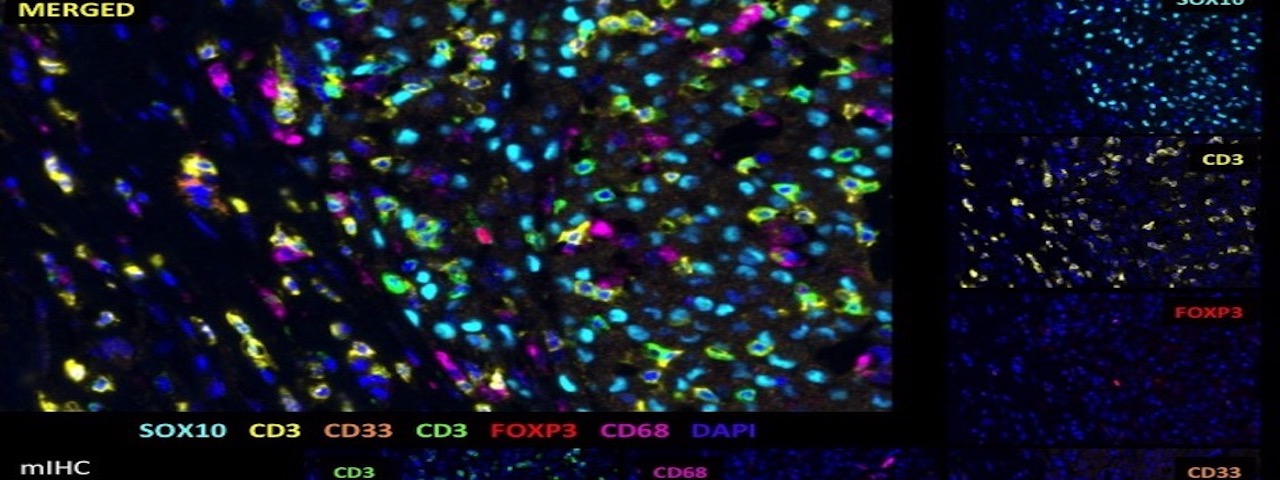
Computational and translational immunology
Cancer immunotherapy has made substantial progress and has dramatically impacted the treatment of multiple cancers. Our group collaborate closely with cancer immunologist and physicians at Moffitt to discover new biomarkers that can better predict the immunotherapy benefits and related outcomes. We defined a new type of eQTL (tis-eQTL) to detect the germline variants underlying the tumor immune gene expressions.
Announcements:
MULTIPLE postdoc fellow positions available at Moffitt Cancer Center (Biostatistics)