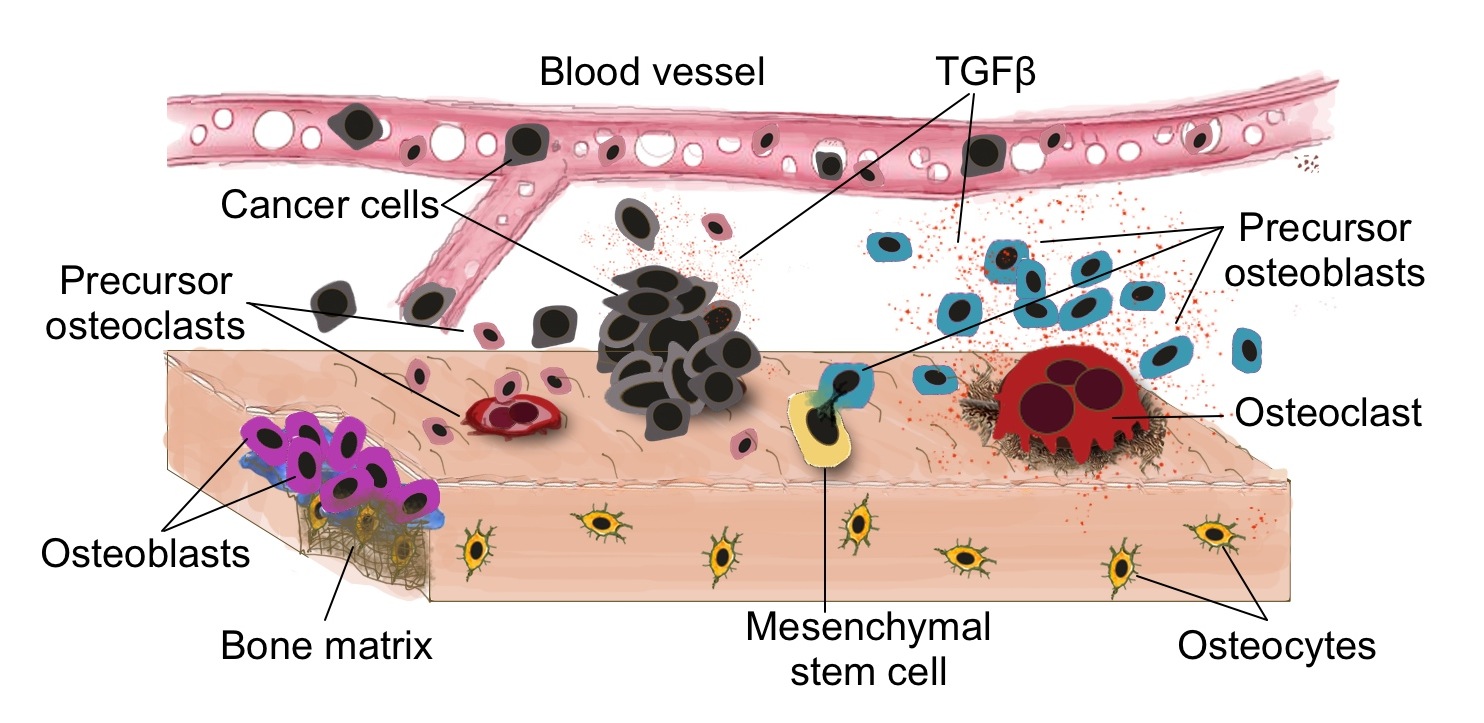
Bone cancer ecosystem (by Arturo Araujo)
We study the ecology of cancer and its evolutionary dynamics and we do that through mathematical tools. Both cancer evolution and intra-tumor heterogeneity are unquestionably key drivers of cancer progression. We concentrate on modeling the roles of the interactions between cancer cells and other cells and between cancer cells and the physical microenvironment in order to characterize the tumor ecosystem and predict cancer evolution.
Game Theory, the brainchild of Janos Von Neumann, was initially used (and still is) to produce models of economics and social sciences, but thanks to the pioneering work of theoretical biologists like John Maynard Smith, it is now one tool widely used by biologists to study evolution within a species in an ecosystem. Once the interactions between the relevant phenotypes (or strategies in game theoretical parlance) have been described, it is possible to deduce both evolutionary stable strategies (ESS) and replicator dynamics. The former will tell us whether a given composition of the population is evolutionary stable, that is, will be able to withstand an invasion from individuals sporting a different phenotype/strategy. The latter will also give us how the composition of the population changes over time until it reaches (if there is) an equilibrium.
Since tumour cells are known to acquire a number of different phenotypes in the path from cancer initiation to malignancy (Hanahan and Weinberg, Cell, 2000), evolutionary game theory can be a powerful tool in which to study the emergence of different tumour phenotypes with increasing degrees of malignancy, the scenarios that lead to benign tumours and the effects of therapies on tumour progression dynamics.
Using Cellular Automata to study the effect of the microenvironment in carcinogenesis “Mathematical modeling in cancer should, therefore, attempt to generalize the extensive data into a single broad paradigm that is internally consistent and sufficiently robust to provide testable predictions for future research” (Gatenby R. A., 1998. Mathematical models of tumour-host interactions. Cancer J. 11 (6), 289293)
We can look at process of cancer initiation and progression (carcinogenesis) from different levels. At the genetic level, the process of DNA replication is not 100% reliable and mutations occur some of which can lead to healthy cells duplicating at a faster rate that they should, and in more advances stages of carcinogenesis, to cells that can degrade the tissue in which they reside, that can invade other tissues or metastasise to other parts of the organism. The number of mutations needed to develop a cancer or the nature of this mutations is still under research and varies heavily from one cancer to another (eg. brain cancer or colon cancer). Alternatively it is possible to look at the different capabilities that tumour cells acquire on their way to make a cancer and study its evolution.
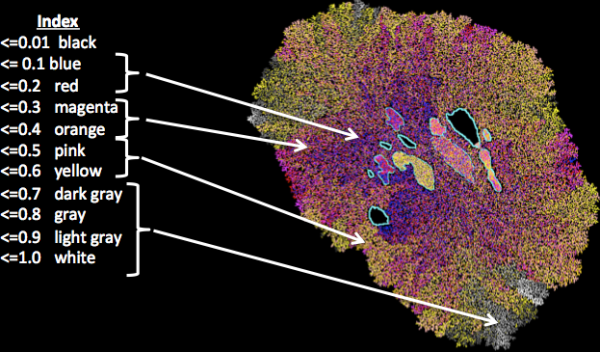
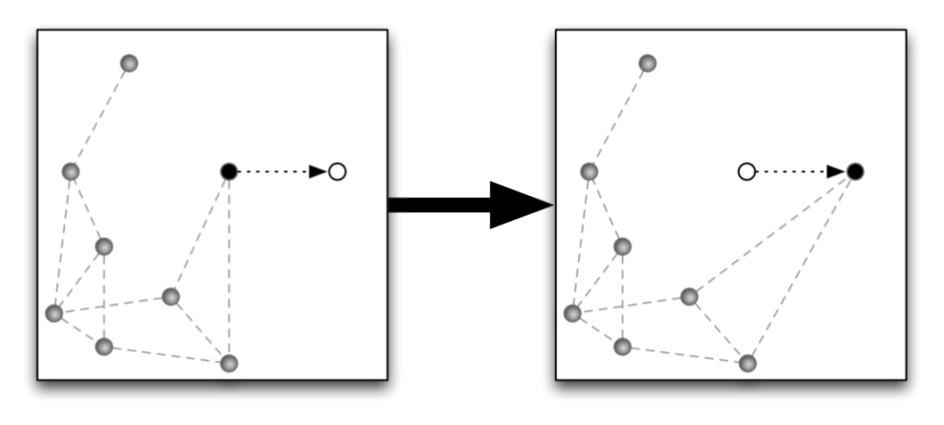
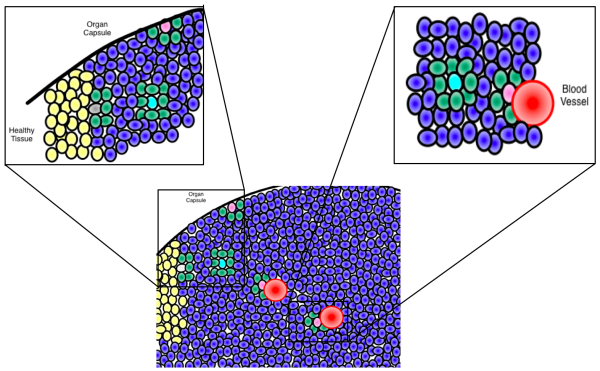
Network theory is often used to understand interactions between proteins or between genes. We look at the cellular scale to see how cells interacting with other cells can predict tumour evolution or even bone structure.