---------- AI 4 DRUG DESIGN ----------
1. Transformer Graph Variational Autoencoder for Molecular Design
Our Transformer Graph Variational Autoencoder (TGVAE) is an innovative generative AI model that employs molecular graphs as input data, thus captures the complex structural relationships within molecules more effectively than string models. To enhance molecular generation capabilities, TGVAE combines a Transformer, Graph Neural Network (GNN), and Variational Autoencoder (VAE). TGVAE outperforms existing approaches, generating a larger collection of diverse molecules and discovering structures that were previously unexplored.
Our goals are to modify TGVAE for more intelligent molecular design, for more specific ligand libraries, and combine structural representation with quantum level interactions description. This advancement not only brings more possibilities for drug discovery but also sets a new level for the use of AI in molecular generation.
Lead Scientist: Trieu Nguyen

2. Mutation-Induced Resistance in Drug Binding Sites:
Integration of Computational Chemistry and Machine Learning
Application of molecular dynamics descriptors as input to the predictive machine learning models presents an innovative way to comprehensively describe protein binding sites and understand the underlying factors of drug resistance. Such integration of computational structural modeling and machine learning with drug design shows promise to transform traditional drug discovery approaches, significantly reducing time and resources while improving the chances of identifying promising drug candidates. Other applications include structure-based drug design and informing our generative AI models.
Lead Scientist: Kat Mizgalska

---------- INTERACTING with INTERACTOMES ----------
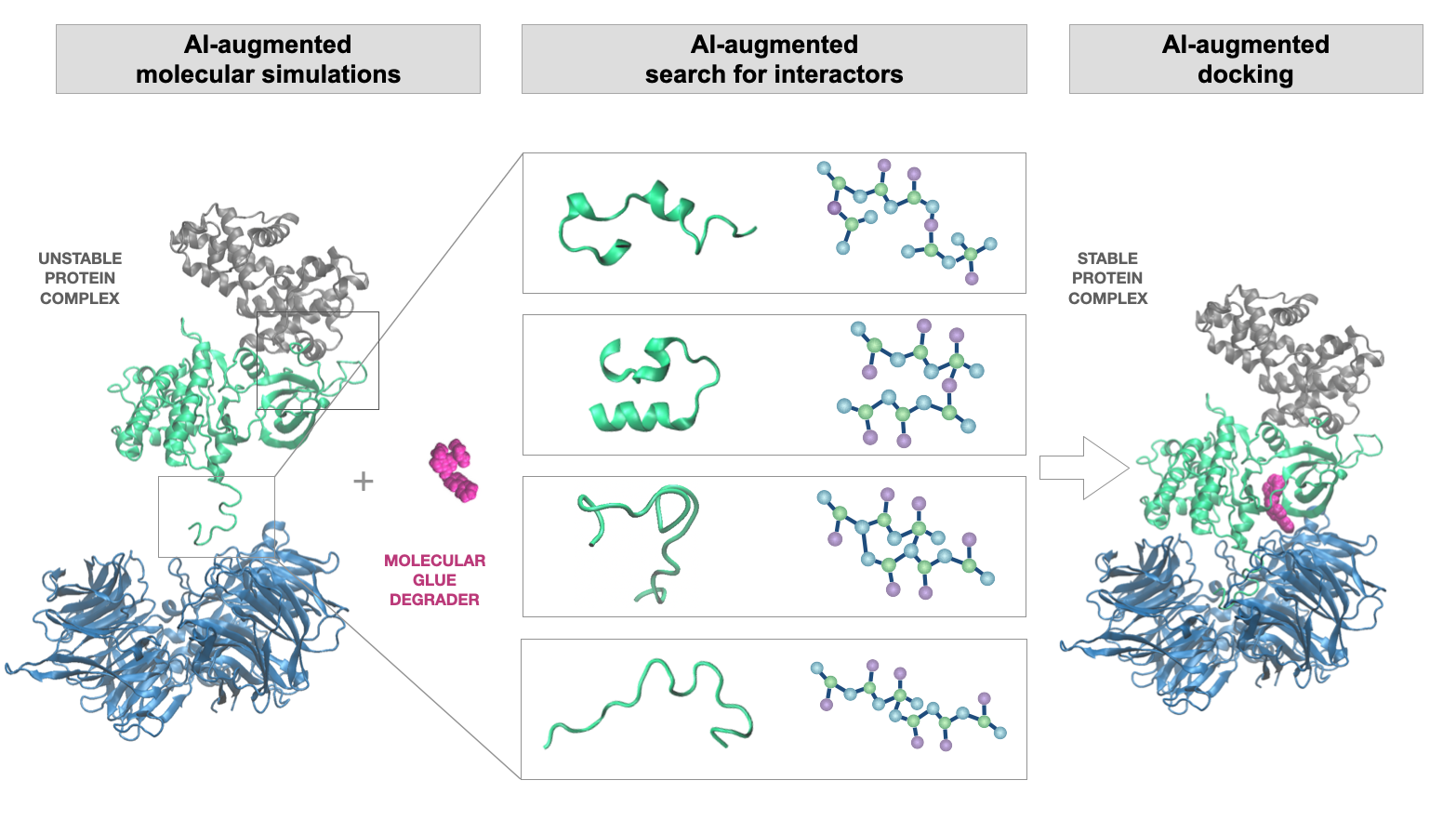
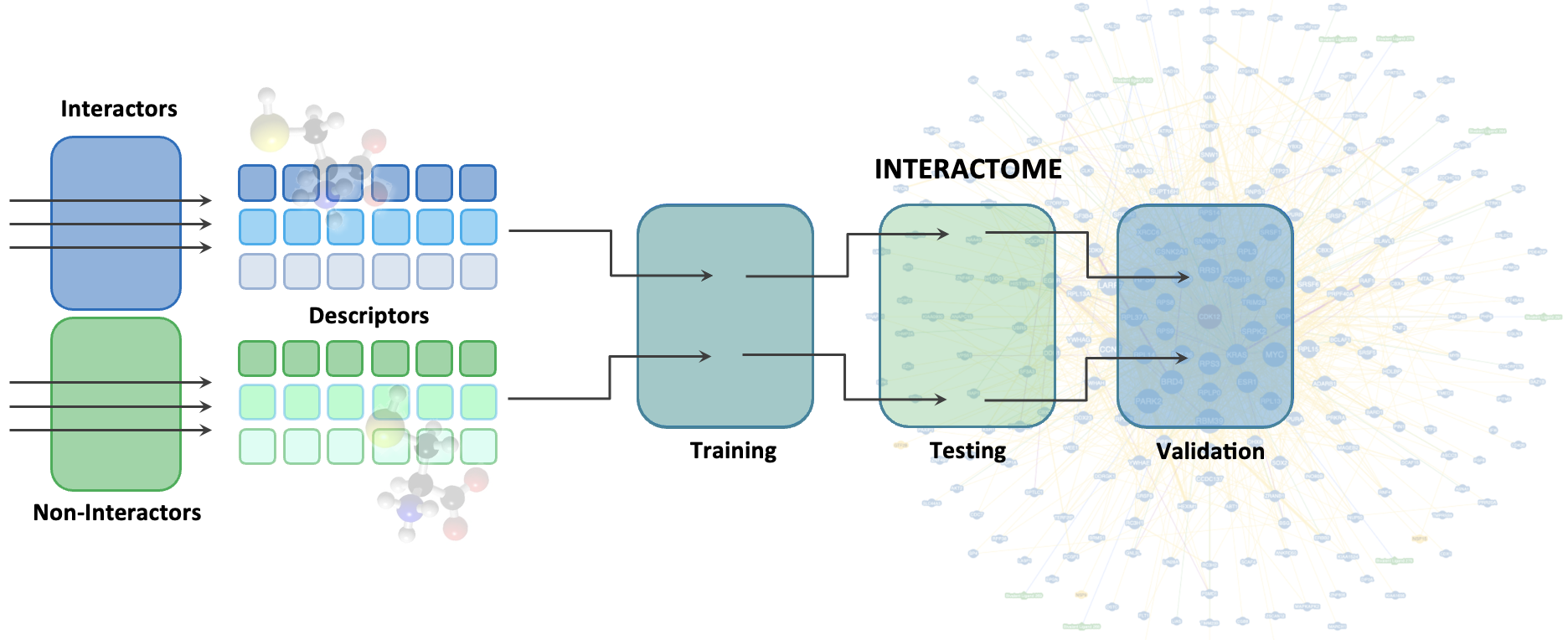
We use AI to augment the existing and develop novel tools for interactive predictions. We utilize computational chemistry (molecular simulations and docking) and machine learning to identify/design and validate the therapeutic candidates that display improved molecular interaction patterns. Besides the structure-dependent binding partner or drug predictions, we also develop the structure-independent models, where numerous physicochemical features allow boost model's performance and accuracy of predictions.
1. Structure-dependent predictions
2. Structure-independent predictions
---------- Genome Organization and Mutagenesis ----------
1. From Nucleotide to Codon to Protein Alterations
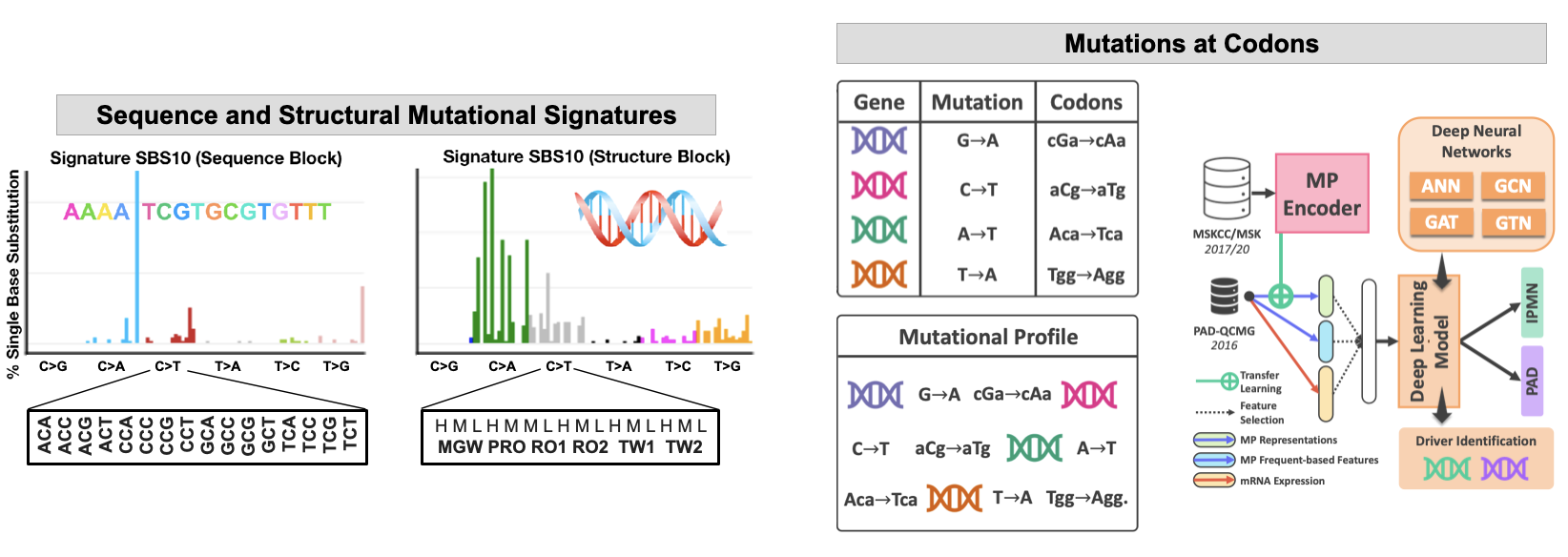
Traditional mutational analyses may not capture all perturbations to the key pathways, especially where large number of genes are involved. We aim to advance pre-malignancy and malignancy classification by developing computational tools that use novel, e.g., structural (conformational) descriptors of the human genome. While exploring DNA spatial organization, we demonstrated that the structural DNA parameters are better predictors of DNA mutagenesis than DNA sequence. We aim to use information about mutations at DNA sequence (3nt or codon) and DNA shape (5nt) to better classify cancers and their subtypes with a goal to predict therapeutic response. Some of our tools include hybrid ML - NLP approach to the mutational data analyses where we derived the text-based information from the mutational profiles of patients’ genomes using the NLP and advanced ML encoders. We further integrate the mutational profiles with codon alterations to guide future analyses of the structural consequences of the mutations on the key proteins and improved drug design.
2. Non-canonical DNA, Mutagenesis, and Drug Resistance
Non-canonical DNA structures detected along gene promoters and other coding regions during transcription and/or replication suggest their importance and relevance for cancer development. During DNA replication, the switch from error-free to error-prone specialized polymerases helps overcome the risk of stopping replication at G-quadruplexes and creates the primary source to acquire mutations. Furthermore, bypassing damaged or non-canonical DNA is recognized as a major mutagen in many cancer types that may lead to drug resistance. Our group is interested in detection of the overlapping non-canonical secondary structures along the genetic material including but not limited to G-quadruplexes and R-loops (DNA-RNA hybrids). Despite their mutagenic potential, these DNA structures can serve as therapeutic targets and lead to cancer suppression.

